-Search query
-Search result
Showing all 30 items for (author: de & la & pena & ah)

EMDB-14784: 
AMC009 SOSIPv5.2 + ACS110 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14785: 
AMC009 SOSIPv.52 in complex with ACS114 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14786: 
AMC009 SOSIPv5.2 in complex with ACS117 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14787: 
AMC009 SOSIPv5.2 in complex with ACS122 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14788: 
AMC009 SOSIPv5.2 in complex with ACS125 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14789: 
AMC009 SOSIPv5.2 in complex with ACS131 Fab
Method: single particle / : van Schooten J, Ward A

EMDB-14783: 
AMC009 SOSIPv5.2 in complex with Fabs ACS101 and ACS124
Method: single particle / : van Schooten J, Ozorowski G, Ward A

EMDB-25634: 
Negative stain map of monoclonal Fab 047-09 4F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25635: 
Negative stain map of monoclonal Fab 241 IgA 2F04 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25636: 
Negative stain map of polyclonal Fab 236.7 binding the anchor and esterase epitopes of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25637: 
Negative stain map of polyclonal Fab 236.7 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25638: 
Negative stain map of polyclonal Fab 236.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
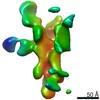
EMDB-25639: 
Negative stain map of polyclonal Fab 236.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25640: 
Negative stain map of polycolonal Fab 236.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25641: 
Negative stain map of polyclonal Fab 236.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
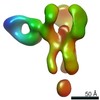
EMDB-25642: 
Negative stain map of polyclonal Fab 241.7 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
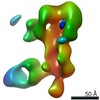
EMDB-25643: 
Negative stain map of polyclonal Fab 241.14 binding the anchor epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB
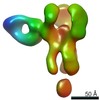
EMDB-25644: 
Negative stain map of polyclonal Fab 241.14 binding the esterase epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25645: 
Negative stain map of polyclonal Fab 241.14 binding an epitope on the top of the head of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25646: 
Negative stain map of polyclonal Fab 241.14 binding the RBS epitope of H1 HA
Method: single particle / : Han J, Richey ST, Ward AB

EMDB-25655: 
CryoEM map of anchor 222-1C06 Fab and lateral patch 2B05 Fab binding H1 HA
Method: single particle / : Han J, Ward AB

EMDB-24720: 
SaPIbov5 procapsid structure including size redirecting protein Ccm
Method: single particle / : Hawkins NC, Kizziah JL, Dokland T

EMDB-22259: 
Human 20S proteasome bound to an engineered 11S (PA26) activator
Method: single particle / : de la Pena AH, Opoku-Nsiah KA, Williams SK, Chopra N, Sali A, Gestwicki JE, Lander GC

PDB-6xmj: 
Human 20S proteasome bound to an engineered 11S (PA26) activator
Method: single particle / : de la Pena AH, Opoku-Nsiah KA, Williams SK, Chopra N, Sali A, Gestwicki JE, Lander GC

EMDB-4590: 
Leishmania tarentolae proteasome 20S subunit complexed with GSK3494245
Method: single particle / : Rowland P, Goswami P

EMDB-4591: 
Leishmania tarentolae proteasome 20S subunit apo structure
Method: single particle / : Goswami P, Rowland P

EMDB-9042: 
Yeast 26S proteasome bound to ubiquitinated substrate (1D* motor state)
Method: single particle / : de la Pena AH, Goodall EA

EMDB-9043: 
Yeast 26S proteasome bound to ubiquitinated substrate (5D motor state)
Method: single particle / : de la Pena AH, Goodall EA

EMDB-9044: 
Yeast 26S proteasome bound to ubiquitinated substrate (5T motor state)
Method: single particle / : de la Pena AH, Goodall EA

EMDB-9045: 
Yeast 26S proteasome bound to ubiquitinated substrate (4D motor state)
Method: single particle / : de la Pena AH, Goodall EA, Gates SN, Lander GC, Martin A
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
